Ordered by weight (if set) and creation date.
Research
My associates and I are using physical biochemical approaches to study what might be called the molecular basis of gene expression. Most of our experimental work is concerned with the function and regulation of the complexes that control DNA transcription and replication, with studies focused primarily on transcription with the E. coli DNA-dependent RNA polymerase and its regulatory factors and on replication with the seven-protein bacteriophage T4-coded DNA replication system. Comparative studies are also underway using selected components of some equivalent eukaryotic systems.
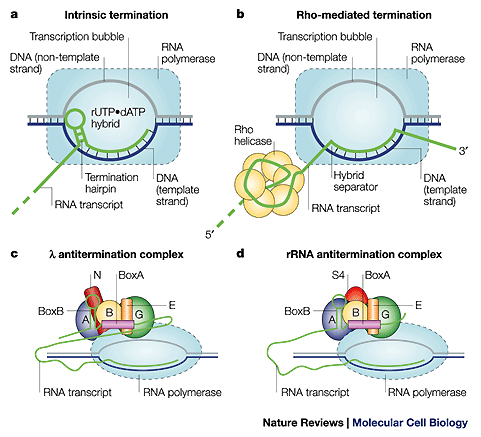
In transcription our group is studying the transcription cycle, both at the overall operon level and at the level of the various steps of the single-nucleotide addition-excision cycle. At the operon level we are studying regulatory interactions that control activation and repression at initiation, the kinetics of elongation, and the molecular bases of the elongation-termination decision at both intrinsic and rho-dependent transcription terminators, together with the mechanisms of action antitermination factors. We have recently completed a study of E. coli transcription termination factor rho, in terms of its function as a specific RNA-DNA helicase and as a transcript terminator. Our antitermination studies have focussed on the mechanisms of action of the N protein of phage lambda in N-dependent antitermination systems. At the single-nucleotide addition-excision cycle level we are using various kinetic techniques to understand the molecular origins of transcriptional processivity and fidelity.
In replication our work began with studies of the cooperative binding of the T4 gene 32 (single-stranded DNA binding) protein to the single-stranded DNA (and RNA). This then led us to examine the interactions of the other components of the system, including those of the DNA polymerase with the primer-template and the polymerase accessory proteins. These studies have shown that the basically nonprocessive T4 DNA polymerase can be rendered fully processive by means of "sliding-clamp" processivity factor, and that the role of the other accessory proteins is to carry out the specific and ATP-dependent loading of the processivity factor onto the polymerase at the primer-template junction in the replication fork. The resulting complex can carry out leading strand DNA replication with essentially in vivo rate, fidelity, and processivity. The helicase of the T4 DNA replication system functions as a hexamer and, in combination with a single T4 primase subunit, forms a stable primosome subassembly. We have recently also shown a direct coupling between the polymerase and the helicase. This complex alone, if properly loaded onto a model replication fork, can carry out processive synthesis on a double-stranded DNA construct at the physiological rate. The mode of assembly of these components into a fully functional and coupled DNA replication system is currently being studied.
In all these studies we emphasize the elucidation of the detailed mechanisms and general principles of protein-nucleic acid and protein-protein interactions that underlie the function of these biologically central complexes.
Termination and antitermination complexes that are being studied in the von Hippel laboratory.
| Attachment | Size |
|---|---|
| vonhippelfig.gif | 48.77 KB |